Connecting to Beckman Biomek#
This guide will walk you through how to connect Artificial to Biomek Software using the Biomek Resource Library.
Supported Beckman Biomek versions#
Beckman Biomek 5.1.10
Install the driver#
Download and install the Artificial Beckman Biomek Driver on the PC where the Biomek software is running.
Note
For more information on how to install and configure the Driver, see our guide on Installing and Configuring Device Drivers.
Install the Resource Library in your Adapter#
Run the following command in the terminal of your adapter repository to install Artificial’s Beckman Biomek Resource Library:
uv add artificial-biomek-resource-library==1.1.*
Your pyproject.toml file should now include artificial-biomek-resource-library in the project dependencies section:
[project]
dependencies = [
...
"artificial-biomek-resource-library==1.1.*"
]
Update your Adapter Config#
config.yaml#adapter:
name: BiomekTutorialAdapter
remoteConfig: False
allowAnySequenceId: True # Useful when running in a local dev container
plugin: # all resources this adapter can connect to
resource:
name: "Biomek i7" # user friendly name, unique in the adapter
id: "biomek" # This should match the device key in the asset_sync section below
driver:
name: "biomek" # Driver name, this is a non-configurable string that needs to match the driver identity
url: "http://biomek.webaddress.com:49835" # URL of the hardware and driver
resource_simulation: false # Set to true to run simulation without hardware
driver_simulation: false # Set to true to run simulation without a driver
cert_file: "adapter/certs/ca.crt"
workspace_name: "C:/biomek_files/Biomek i7.bif" # Modify to use desired workspace name
asset_sync:
devices: # device string names/prefix must match resource id above
"biomek": { rid: "d1234567-34d0-4391-be64-7aef4e0b28be" }
Fill in the correct URL (including the port) for the Biomek device. The device’s IPv4 URL can be obtained from Network & Internet Settings or by running
ipconfigin a command window. The port is listed in the running driver server console window.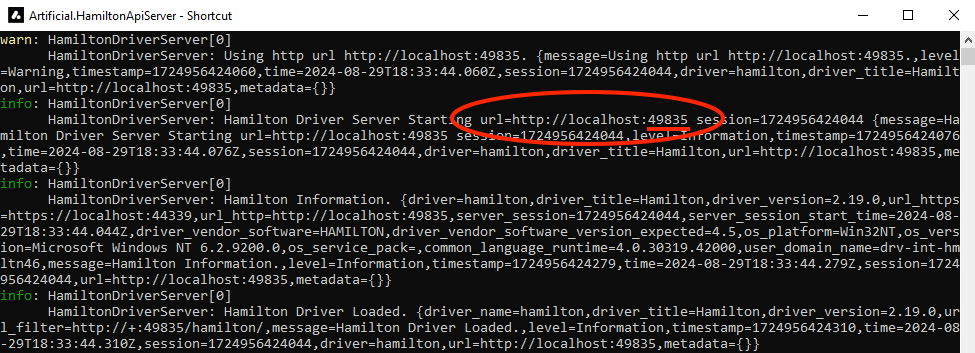
Fill in the workspace path to match the installation location on the Biomek device.
You may update the resource name above if you wish. Any string will work but it must be unique across the instance.
If you wish to run in simulation without hardware or without a driver, change
resource_simulationordriver_simulationtotrue, respectively.Update the
asset_syncsection to map the resource id to the instrument’s asset instance id in the digital twin. You can fill out the rest of the devices later. See Asset Sync Config for more information.
Use the Resource Library in your Adapter#
adapter/main/plugin.py file#from artificial.adapter_common import ActionModulePlugin, ability_name, action_modules
from artificial.adapter_common.plugin import PluginContext, plugin_config
from artificial.biomek.actions import BiomekActions
from artificial.biomek.core import BiomekResource
from artificial.biomek.event_handler import BiomekEventHandler
from artificial.biomek.models import PluginConfig
from artificial.logging import get_logger
from artificial.resource_base.asset_syncer import ResourceAssetSyncer
from artificial.resource_base.models import SyncConfig
# from .simulators.labware_simulator import BiomekDriverLabwareMock as LabwareSimulator
logger = get_logger(__name__)
@ability_name('biomek')
@action_modules(BiomekActions)
class AdapterPlugin(ActionModulePlugin):
"""assemble resources components with resource configuration"""
_cfg = plugin_config(PluginConfig) # this will make PluginConfig show up in the adapter config UI
async def setup(self, pctx: PluginContext) -> None:
prog_config = pctx.config
plugin_conf = self._cfg
sync_config: SyncConfig = pctx.raw_config.to_dataclass(SyncConfig, 'adapter.asset_sync')
logger.debug(f'sync_config loaded: {sync_config}')
# all resources in the adapter use the same res_syncer
syncer = pctx.asset_sync_manager_v1(f'ResourceSyncer-{prog_config.adapter.name}')
res_syncer = ResourceAssetSyncer(syncer, sync_config)
await res_syncer.initialize()
# create instances of resources for this adapter
resource = BiomekResource(
pctx.alabPyBase,
lab_id=prog_config.artificial.labId,
adapter_id=prog_config.adapter.name,
resource_id=plugin_conf.resource.id,
name=plugin_conf.resource.name,
res_syncer=res_syncer,
)
if resource:
resource.set_driver(driver_config=plugin_conf.resource.driver, simulator=None)
await resource.set_health_monitors(pctx.lab.health)
module = BiomekActions(resource)
self.add_module(module)
# subscribe the driver events
event_handler = BiomekEventHandler(resource)
resource.add_event_handler(event_handler)
Add the required Actors#
Add the following actors to the list of actors in adapter/main/__main__.py.
actors = [
ActorConfig(id='biomek', abilities={'run_protocol': 1, 'substrate': 1, 'biomek': 1}),
]
Publish and run a test Workflow#
This workflow will run a method of your choosing and log a message in the UI upon successful completion. You will define which method to run in the request UI by inputting the method name (e.g., mymethod).
Note
Update the variable params to reflect the needs of your protocol. If your protocol does not require variables, set params to an empty list ([]).
from typing import List
from artificial.workflows.decorators import parameter, workflow
from artificial.workflows.runtime import show_info
from stubs.stubs_actions import VariableType, run_protocol_to_completion
@workflow('Simple Biomek Connectivity Test Workflow', 'simple_biomek_connectivity_test_workflow')
@parameter('biomek_method_name', {'required': True, 'uiTitle': 'Biomek Method Name'})
async def simple_biomek_connectivity_test_workflow(biomek_method_name: str = 'alab-test') -> None:
params: List[VariableType] = [
VariableType(var_name='run_now', var_value='true', var_type='TrueFalse'),
VariableType(var_name='nplates', var_value='2', var_type='Numeric'),
]
await run_protocol_to_completion(biomek_method_name, params) # when no path specified, driver's workspace is used
await show_info('Congratulations, you successfully ran your hardware!', 'Hardware Success')